Guide to the Application of Genotyping to Tuberculosis Prevention and Control
Applying Genotyping Results to Tuberculosis Control Practices
Evaluating Matching Genotypes
The first step in responding to new genotyping results is to identify any new genotyping matches contained in the laboratory report. The genotyping laboratory will flag all isolates with matching spoligotypes and MIRU types by assigning them a PCR cluster designation. Except in rare instances, two persons who are involved in the same chain of recent transmission will have isolates with matching genotypes; conversely, two persons with nonmatching isolates are rarely involved in the same chain of transmission (See Chapter 4, Combining Genotyping and Epidemiologic Data to Improve Our Understanding of Tuberculosis Transmission, for an explanation of the rare exceptions to these rules).
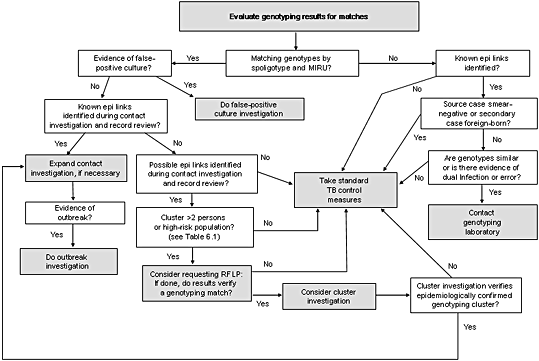
Figure 6.1. Flow diagram describing the evaluation of genotyping results. This diagram describes the action steps (shown in grey boxes) that might result from an analysis of new genotyping data.
Could Genotyping Matches be Due to False-Positive Cultures?
When new genotyping matches are reported, the first question to pose is whether one or more of the matching isolates might represent false-positive cultures? Since the discovery of a false-positive culture may require that a patient’s treatment be stopped, the TB program must maintain a high index of suspicion for the possibility of false-positive cultures, and there should be no delay between receiving new genotyping results and determining if any matches might need further investigation.
False-positive cultures occur when M. tuberculosis bacteria from one specimen, instrument, or culture inadvertently contaminate another specimen or culture, when clerical errors occur and specimens are mislabeled or misreported, or when data entry errors occur. Clinical equipment (e.g., bronchoscopes, sputum collection booths, and ultrasonic nebulizers), if inadequately cleaned, can become contaminated and be the source of false-positive cultures (as well as the source of nosocomial transmission). Cross contamination can occur in the laboratory during batch processing, pipetting, transfer of bacilli from a broth-culture system, work in a faulty exhaust hood, and species-identification procedures.
One of the most important advantages of routinely fingerprinting all M. tuberculosis isolates is the ability to establish an early warning system to identify suspected false-positive cultures. Although this discussion is based on the assumption that the suspected false-positive culture was identified through a genotyping laboratory report, this suspicion can also be raised by health-care providers who receive a questionable culture result from a clinical laboratory, from the laboratory itself when more than one M. tuberculosis culture that was processed during the same time period become positive, or by health department staff members who investigate TB cases.
The following text box lists the factors to look for in evaluating whether a cluster might involve one or more false-positive culture results. If any one of the factors listed in the text box are identified, a false-positive culture investigation should be conducted immediately (see Suspected False-Positive Culture Investigations later in this chapter for details). Note that to evaluate the factors listed, the TB program needs access to data about how and when the specimens were processed and whether particular patients have only one or more than one positive culture result. If these data are not readily available at the time the genotyping matches are reported, the TB program should update their data collection and management procedures.
If this initial evaluation determines that the genotyping matches do not represent a false-positive culture, the next step in the decision analysis depicted in Figure 6.1 is to decide if patients in the cluster share known epidemiologic links.
Summing Up: False-Positive CulturesSuspect that a genotyping match might represent a false-positive culture if any of the following are true:
|
Action Steps for Genotyping Clusters with Known Epidemiologic Links Identified
Known epidemiologic links (see Chapter 4,Combining Genotyping and Epidemiologic Data to Improve Our Understanding of Tuberculosis Transmission, for definitions of epidemiologic links) between at least some of the members of the same genotyping cluster suggest that they are part of the same chain of recent TB transmission. The evidence in favor of recent transmission is strong enough at this point in the decision analysis that it is usually not necessary to collect additional data by requesting RFLP analyses of the isolates or by conducting a cluster investigation for the purpose of deciding on appropriate action (see Chapter 5, Developing a Tuberculosis Genotyping Program, for exceptions to this rule).
Two possible action steps are described in this situation in Figure 6.1: a) expanding the contact investigations or b) initiating (or expanding) an outbreak investigation. The decision about which step to take is based on what is known about the cluster from previous investigations and whether the cluster of cases has grown to be an outbreak.
Deciding when an epidemiologically conformed genotyping cluster has become an outbreak is always a challenge. The following text lists characteristics of outbreaks that may help in this decision-making process; an outbreak is likely occurring if any one of the following criteria is met.
Three criteria for defining an outbreak
1. An increase in the expected number of TB cases.
Although this is a standard epidemiologic definition, it is at times a challenge to apply, since the expected number of TB cases in a specific setting is often difficult to know. This criterion is more useful if it is applied to specific subgroups of persons who share demographic characteristics or specific exposures rather than to the population as a whole.
For example, two unrelated children with TB who go to the same school and are in the same classroom would arguably meet the “unexpected increase” criterion for an outbreak, since under normal classroom circumstances the expected number of children with TB would be zero. Two homeless men with TB in the same city, on the other hand, would probably not meet this criterion, since TB in homeless men is more common than in school-age children.
Universal genotyping will help considerably in identifying outbreaks, since it will help to provide an answer not only to the question “is there an increase in the number of cases in a subgroup?” but also to the question “is there an increase in cases that belong to a specific genotyping cluster that involves recent transmission?” Another helpful aspect of having genotyping results available is that they will help define the scope of an outbreak, since genotyping results can identify persons with genotyping links to outbreak cases where no epidemiologic links were known.
2. Transmission continues despite adequate control efforts by the TB program.
For example, two TB patients who work at the same job might not be considered an outbreak. If each was well investigated, including thorough contact investigations with appropriate screening and treatment for LTBI, there might be little else to do. If, however, an additional person with TB appeared a month later with an isolate that was a genotype match to the isolates from the other two persons, this might meet the criterion of ongoing transmission despite an apparently adequate TB-program response.
3. The contact investigation has grown to a size that requires additional outside help.
This often happens when a TB program is devoting most of its resources to conducting a large contact investigation, but the demand for resources continues to increase to the point that the program cannot meet its routine obligations for basic TB control practices. At this point, declaring the situation to be an outbreak can lead to obtaining additional resources to conduct an outbreak investigation from the state program or from CDC. If the decision is made to conduct an expanded contact investigation or an outbreak investigation, see the respective sections for guidance about how to proceed.
Genotyping Clusters with Possible Epidemiologic Links Identified
The next decision point shown in Figure 6.1 focuses on genotyping clusters where known epidemiologic links have not been established. The next question to consider is whether previously collected data from contact investigations and record reviews show that persons in a genotyping cluster share possible epidemiologic links. If possible epidemiologic links are identified (e.g., the TB patients live in the same neighborhood or were homeless) (see Chapter 4,Combining Genotyping and Epidemiologic Data to Improve Our Understanding of Tuberculosis Transmission, for a definition of possible epidemiologic links), the patients might be involved in the same chain of recent transmission and further investigation might be helpful, especially if the genotyping cluster has more than two patients or if the patients are considered high-risk. If the RFLP results show that the presumed cluster is, in fact, made up of genetically distinct isolates (i.e., the isolates have non-matching RFLP patterns), there is no need to conduct a cluster investigation.
If, on the other hand, the RFLP results confirm that the isolates belong to the same PCR/RFLP cluster (i.e., if the RFLP patterns match), the TB program should consider conducting a cluster investigation
Whether to Launch a Cluster Investigation
Cluster investigations should be launched only when needed, since they can be labor intensive. If RFLP results confirm that the isolates belong to a single PCR/RFLP cluster, it is helpful to compare the characteristics of the cluster with the prioritization scheme described in Table 6.1. This prioritized list should not be interpreted as providing absolute instructions about when to conduct a cluster investigation and when not to; rather, it provides helpful guidance about when a cluster investigation is needed and when it might be wise to wait to see if additional TB patients are identified as belonging to the cluster. The information that this prioritization scheme is based on comes from the NTGSN study, and parts of it may have to be updated when our experience with the PCR genotyping methods grows.
Most commonly, clusters involve only two persons. One way to save valuable TB program resources is to investigate only clusters that involve at least three persons. Some programs will want to investigate clusters of only two persons; if resources are available to do so, this aggressive approach will identify episodes of recent transmission sooner. If resources are scarce, however, conducting investigations only of clusters with at least three persons is a reasonable policy. In fact, if resources are really scarce, conducting a cluster investigation only when the cluster grows to four persons may have to be adopted as an interim policy until more resources are identified.
The decision as to what cluster size should be investigated is influenced by whether the cluster contains high-risk persons. “High-risk” in this setting refers to a) characteristics of the persons in the cluster that might make them particularly infectious (e.g., having cavities on chest radiographs), b) characteristics of the M. tuberculosis strain that make it particularly dangerous (e.g., a multidrug-resistant strain), or c) characteristics of persons in the cluster that, if shared by their exposed contacts, would increase the risk of progression from LTBI to active TB (e.g., HIV infection or other immunocompromising conditions) or would increase transmission among a group (e.g., jail inmates, nursing home residents, the homeless). A TB program that would otherwise decide to conduct investigations only of clusters containing at least three persons might decide to conduct an investigation of a two-person cluster if one or the other persons with TB was considered “high-risk.”
Additional information that comes from future genotyping laboratory reports may tip the balance in favor of conducting an investigation. If one laboratory report identifies a two-person cluster, the TB program might decide not to begin a cluster investigation if neither of the two persons is “high-risk.” If the next laboratory report identifies a third person with a matching genotype, the TB program will probably want to initiate a cluster investigation of all involved persons. In general, the decision whether to launch a cluster investigation is a dynamic process, and a decision at an early stage not to do one should not inhibit a TB program from changing its mind should new information become available. If the decision is made to conduct a cluster investigation, details on how to do so are provided later in this chapter in the section titled, Cluster Investigations.
Table 6.1. Prioritizing genotyping cluster investigations.
| Priority (from high to low) |
Type of cluster | Rationale for priority |
|---|---|---|
|
|
Suspected false-positive culture | Need to determine which patients do not have TB and stop treatment |
| Cluster of three or more high-risk* patients with possible epidemiologic links | Need to confirm or exclude recent transmission in large clusters of high-risk* patients | |
| Cluster of two high-risk* TB patients with possible epidemiologic links | Smaller clusters less likely to yield epidemiologic links, but presence of high-risk patients deserves attention | |
| Cluster of three or more low-risk TB patients with possible epidemiologic links | Investigation of low-risk patients less urgent than high-risk* patients, but larger clusters may deserve attention | |
| Cluster of two low-risk TB patients with possible epidemiologic links | Investigation of smaller clusters of low-risk patients often does not yield helpful information Investigations can, however, provide data for monitoring program performance |
|
| Cluster of high-risk* TB patients who have not been found to have even possible epidemiologic links | Low yield for establishing new epidemiologic linksInvestigations can, however, provide data for monitoring program performance Reserved for programs with sufficient resources | |
| Cluster of low-risk TB patients who have not been found to have even possible epidemiologic links | Very low yield for establishing new epidemiologic links Investigations can, however, provide data for monitoring program performanceReserved for programs with sufficient resources |
* "High risk" is defined as patients living in congregate settings (e.g., correctional institutions and nursing homes), persons infected with HIV or having other immunocompromising conditions, children, patients with cavities on chest radiographs or with MDR TB, and the homeless.
Genotyping Clusters with No Epidemiologic Links
Not all genotyping clusters represent recent transmission, and conducting a cluster investigation when the chance of gaining new information is slim is often not a wise investment of resources. If information from adequately conducted contact investigations and case interviews does not reveal even possible epidemiologic links between patients in a genotyping cluster, it is probably sufficient for TB programs to simply ensure that standard TB control measures are completed, such as ensuring that all cases are completely treated and all infected contacts are identified and treated appropriately. Of course, if future genotyping matches are identified and new patients are added to a genotyping cluster, new epidemiologic links within the cluster may become apparent, and at that point the TB program may need to initiate a cluster or an outbreak investigation.
- Page last reviewed: September 1, 2012
- Page last updated: September 1, 2012
- Content source:


 ShareCompartir
ShareCompartir