Using whole genome scans to discover obesity genes: Implications for the Clinical Utility of Genetic Testing in Obesity Quix
This website is archived for historical purposes and is no longer being maintained or updated.
Katrina A.B. Goddard
American Society of Human Genetics Fellow
Office of Public Health Genomics
Centers for Disease Control and Prevention
Educational objectives
After reading this case study, you should be able to:
- Evaluate findings from a GWAS approach
- Calculate measures of clinical validity and utility
- Understand the underlying components that contribute to the ‘value added’ of a genetic test
- Discuss potential clinical or public health benefits of genetic testing
Introduction
Approximately 30% of US adults are now obese (Body Mass Index (BMI)≥30 kg/m2) [Hedley et al., 2004], representing a significant health problem in the US and other developed countries. Obesity is associated with considerable morbidity and mortality through its association with type 2 diabetes, heart disease, metabolic syndrome, hypertension, stroke, and cancer. Obesity is commonly measured using the body mass index (BMI) [weight/height2 in kg/m2], although recently alternative measures, such as the waist-to-hip ratio, have been suggested that may be better predictors of mortality [Welborn, 2007]. Although lifestyle factors, such as diet and exercise, are important determinants of obesity, genetic studies have produced estimates of heritability for BMI between 30-70% [Bell et al., 2005; Farooqi et al., 2005; Hebebrand et al., 2003; Schousboe et al., 2003].
Genome-wide Association Studies (GWAS) are an increasingly popular tool to search for genetic risk factors that contribute to susceptibility to disease. The advantages of the approach include the ability to conduct population-based studies, which may be easier and less expensive than family studies. They may also have increased power to detect loci with small effect, a likely circumstance for complex traits, and allow finer localization of the signal. The drawbacks of this approach include sensitivity to alternative causes of allelic association, including population stratification and chance (type II error), as well as reduced power in the presence of locus and allelic heterogeneity or mutations that arise more than once.
Case Study
In April 2006, Herbert et al. reported that a common variant near the insulin-induced gene 2 (INSIG2) is associated with obesity, which was identified through a genome wide association approach. The primary study population was the NHLBI Framingham Heart Study, where individuals were enrolled from the community, and were not selected for a particular trait or disease. Herbert, and coworkers, then genotyped the variant in five additional studies including the Nurses Health Study; the KORA S4 cohort from a town near Munich, Germany; a case-control study of subjects from Poland and the United States, in which cases were selected based on BMI; a sample of African-American families and unrelated individuals from Maywood, Illinois, who were selected based on BMI; and a sample of Western European parent-child trios who were selected because of obesity in the child. Association was detected in four of the five replication studies. The high-risk genotype is present in approximately 10% of the population, and confers a risk approximately 1.22-1.33 times the risk among persons without the high risk genotype.
Herbert A, et al., A Common Genetic Variant is Associated with Adult and Childhood Obesity. Science 312:279-283, 2006.
- What approaches were used to control for alternative explanations of the observed allelic association?
- Family-based test of association
- Correction for multiple testing
- Replication in additional samples
- All of the above
- In this study, 86,604 SNPs were tested for association with BMI, and at least two genetic models were considered, including a recessive mode of inheritance. The most significant SNP, rs7566605, had an unadjusted p-value of 0.0026, which was claimed to reach overall significance. Which method was used to correct for multiple testing, and what threshold was used to determine statistical significance?
- 5.8 x 10-8
- 0.005
- 0.025
- 0.05
- Table 2 shows the location and p-values for the four neighboring SNPs to rs7566605 including one SNP located only 184 bp away. None of these SNPs show strong evidence of association with BMI. Which of the following explanations is a possible interpretation of this finding?
- The association between BMI and rs7566605 is a false positive result.
- The neighboring loci are not good proxies for rs7566605 because the inter-marker LD is low.
- The association between BMI and rs7566605 is a true finding, but association with neighboring markers is not observed because an alternative model, such as recurrent mutation at a functional SNP (rs7566605), is needed to explain the results.
- All of the above
- Several different populations were used to replicate the initial finding from the Framingham Heart Study. What characteristics do you think are important when selecting populations to confirm findings from a GWAS? Were your criteria met in this instance?
- The SNP rs7566605 lies between two genes, FLJ10996 and INSIG2. Do the data presented in this paper answer definitively which gene may be involved in obesity risk?
- Yes
- No
- Assuming the risk of obesity in the population is 30%, a relative risk of 1.33 for the high risk genotype (CC) compared to the low risk genotypes (CG, GG), an allele frequency of 0.373 for the C allele, and Hardy-Weinberg Equilibrium, what is the risk of obesity for individuals with each genotype?
- The risk of obesity is 38% for individuals with the CC genotype
- The risk of obesity is 29% for individuals with the CG or GG genotypes
- Both a and b are correct
- Both a and b are incorrect
- If you were to develop a genetic test to predict individuals at increased risk of developing obesity based on this variant, what would be the sensitivity and specificity of the test?
- The sensitivity is 0.974
- The specificity is 0.879
- Both a and b are correct
- Both a and b are incorrect
- The principal investigator for this study was quoted as saying “There is no immediate benefit to this finding”. Based on the calculations of specificity and sensitivity in the previous question, do you agree with his statement?
- Yes
- No
- In a population of 10,000 individuals, how many cases of obesity are attributable to carrying the high risk genotype?
- 130
- 525
- 1000
- 1390
- The population attributable risk percent (PAR%) for this genotype is 4.33% (this is what is usually referred to as the PAR), and the population attributable risk (PAR) is 1.3% (also called the attributable community risk [ACR]). What does this mean?
- Obesity could be prevented in 4.33% of cases by eliminating the effects of this locus
- Obesity could be prevented in 1.3% of the population by eliminating the effects of this locus
- Both a and b are correct
- Both a and b are incorrect
- Suppose an intervention is developed that eliminates the effects of this locus (i.e., the risk of obesity for CC individuals is reduced to the risk observed for the GG or GC individuals), perhaps a therapy that reduces plasma triglyceride levels, a mechanism suggested in the paper. Since some individuals with the GG and GC genotypes may also have elevated triglyceride levels, such a treatment could plausibly reduce the risk of obesity for them as well. For each scenario, describe the circumstances for which you would recommend
- treating everyone with obesity, or
- screening for high triglycerides prior to treatment, or
- screening for the CC genotype prior to treatment?
- Suppose the intervention in the previous question is equally effective in reducing the risk of obesity in everyone, regardless of genotype (CC, GC or GG), e.g., a weight management program. Some have argued that screening for the CC genotype may still be beneficial if it helps persuade people with the CC genotype to comply with the intervention. One shortcoming of this strategy is that people withoutthe high-risk CC genotype may be less likely to comply because they are convinced they are not at risk.
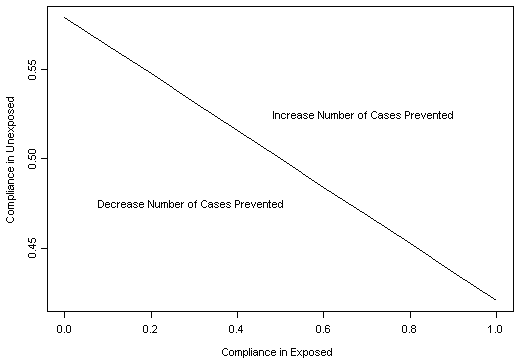
The line in the figure below shows the rates of compliance by genotype--CC (exposed) vs. GC or GG (unexposed)--for which the number of cases prevented by this intervention is equal to a predefined value. Changes in compliance rates in either group will change the number of cases prevented. Assume that compliance with the weight management program prior to genotyping is 50%. If, after genotyping, compliance is increased to 80% in the group with the CC genotype, how much can compliance be reduced for those with the GC or GG genotypes without eliminating the beneficial effect of genotyping?- 3%
- 5%
- 10%
- 30%
- Eight months after the initial discovery was published by Herbert et al., three additional groups wrote letters to the editor reporting findings on the same variant. Loos et al. (2007) evaluated two ethnically homogeneous, population-based cohorts, including 4916 subjects from the EPIC Norfolk study and 1683 subjects from the Medical Research Council (MRC) Ely study. Rosskopf et al. (2007) evaluated a large German population-based, cross-sectional study of 4310 unrelated persons. Dina et al. (2007) evaluated 10,265 persons of French-Caucasian descent, who were recruited for several different studies with family-based, case-control, or general population designs, some of which has recruitment criteria based on obesity phenotypes.
All of the replication studies used the same phenotypic measure (BMI, with those ≥30 kg/m2 considered obese), genotyped the same variant (rs7566605), and examined populations similar to that of the initial study (Caucasian). However, none of the studies found significant evidence of association between genotype and BMI; in fact, there was a slight tendency towards the opposite effect. The only exception was a subgroup analysis of the Rosskopf et al. study, which found a significant positive association between CC genotype and mean BMI among 2701 overweight participants.
In response, Herbert et al. suggested several possible explanations for the discrepancy in findings, including differences in study design, heterogeneity of effect size, and the “winner’s curse”. What is the “winner’s curse”, and do you think this is a plausible explanation for the inconsistency in the study findings?
Correct Answers:
Question 1:
Answer d is correct:
All of the above is the correct answer.- Family-based association tests (FBAT) are one approach that is used to control for spurious associations due to population stratification. The primary analysis for the initial study was an FBAT approach.
- A two-stage procedure and Bonferroni correction were used to correct for multiple testing. In the first stage, ten markers were selected for further analysis. The Bonferroni correction was then applied for the FBAT analysis, which was only conducted for these 10 markers.
- After the initial association was detected, the SNP was genotyped in 5 additional study samples to determine if the original signal could be replicated. Meta analysis was performed to evaluate the evidence of association across all of the study samples.
Question 2:
Answer b is correct:
If a Bonferroni correction were applied for all 86,604 SNPs that were genotyped, a significance level of 5.7 x 10-7 would be necessary for statistically significant evidence of linkage. To avoid the risk of false negatives, because the Bonferroni correction is known to be a conservative approach, a two-stage procedure was used. In the first screening step, markers are selected for further analysis by estimating the genetic effect size. The offspring phenotype is predicted using the expected value of the offspring genotype given the parental genotypes (not the observed offspring genotype). The loci with the highest genetic effect are then evaluated in the test step using the measured offspring genotypes. The supplemental online material shows that the screening step and the test step use statistically independent information. Therefore, the Bonferroni correction was then applied only for the 10 SNPs that were analyzed in the second stage. Thus, a p-value less than .005 (answer b) was considered statistically significant, which was reached by SNP rs7566605 with a p-value of .0026.Question 3:
Answer d is correct:
All of the explanations provided are possible, although the authors suggest b as the explanation for the observations.Question 4:
Ideally, at least one replication study would be conducted in a large sample selected from the same population and using the same selection criteria and phenotype definitions as the original study. This would reduce the possibility of a false negative finding in the replication sample due to genetic heterogeneity. None of the five replication studies met these criteria; however, all of them found a statistically significant association with the variant of interest, which supports the generalizability of the study finding.
Question 5:
Answer b is correct, no:
The linkage disequilibrium surrounding the positive SNP extends into both genes. Although association with the INSIG2 gene is biologically plausible, this does not prove it is the causative gene, particularly given the relative lack of information available about FLJ10996. The best evidence would be to show that one of these genes has a functional mutation and demonstrating how it caused obesity.Question 6:
Answer c is correct:
a and b are correct.Question 7:
Answer b is correct:
The correct value for sensitivity is .174.Question 8:
either answer is correct (an opinion):
However, typically a greater sensitivity and specificity is desired for a predictive test.Question 9:
Answer a is correct:
For a nice explanation of the difference between population attributable risk and attributable community risk (which was asked for here) see Wacholder, Epidemiol 16 (1):1-3, 2005, with a comment in Wacholder, Epidemiol 16(4):594, 2005.Question 10:
Answer c is correct:
Both a and b are correct.Question 11:
- Treating everyone with obesity. This strategy might be suggested if the cost of testing for high triglycerides is high compared with the cost of treatment, and the risks of treatment for individuals without high triglycerides are low (e.g., a dietary intervention).
- Screening for high triglycerides prior to treatment. This strategy might be suggested if the treatment is effective for everyone with high triglycerides, and there is no additional benefit for individuals with the high risk genotype (e.g., increased motivation or efficacy of treatment).
- Screening for the CC genotype prior to treatment. This strategy might be suggested if the treatment has greater efficacy for those with the CC genotype, and treatment costs would be too high compared to the benefit for individuals with high triglycerides, but without the CC genotype (potential adverse reaction, or reduced efficacy in this group).
Question 12:
Answer b is correct:
The relative values of the change in compliance for those with CC genotype compared with the change in compliance for those with GG or CG genotype depends on the frequency of the high risk genotype. In this case, since the CC genotype has a relatively low frequency (~10%), a large change in compliance for this group can easily be offset by a small change in compliance for the much larger group with GG or CG genotypes.Question 13:
The “winner’s curse” occurs when there are multiple causes of a disease (e.g., locus heterogeneity), but because of sampling variability, the first study to describe an association has more than the expected number of persons with one of the many potentially causal genetic variants. This variant is not over-represented in subsequent studies; thus, larger sample sizes are typically necessary to replicate the initial finding. This does not seem like a plausible explanation for the discrepancy in these results because the replication studies were much larger than the initial study (which included only 694 persons); furthermore, the frequency of the high risk genotype was similar among studies.
References
- What approaches were used to control for alternative explanations of the observed allelic association?
- A. A. Hedley et al., JAMA 291, 2847 (2004).
- C. G. Bell, A. J. Walley, P. Froguel, Nat. Rev. Genet. 6, 221 (2005).
- I. S. Farooqi, S. O'Rahilly, Int. J. Obes. 29, 1149 (2005).
- J. Hebebrand, S. Friedel, N. Schauble, F. Geller, A. Hinney, Obes. Rev. 4, 139 (2003).
- K. Schousboe et al., Twin Res. 6, 409 (2003).
- C. Dina et al., Science 315, 187b (2007).
- R. J. F. Loos et al., Science 315, 187c (2007).
- D. Rosskopf et al., Science 315, 187d (2007).
- A. Herbert et al., Science 315, 187e (2007).
- Page last reviewed: June 15, 2009 (archived document)
- Content source:


 ShareCompartir
ShareCompartir