Outbreak of Influenza A(H3N2) Variant Virus Infections Among Persons Attending Agricultural Fairs Housing Infected Swine — Michigan and Ohio, July–August 2016
Weekly / October 28, 2016 / 65(42);1157–1160
Rebekah S. Schicker, MSN, MPH1,2; John Rossow2,3; Seth Eckel, MPH4; Nicolas Fisher5; Sally Bidol, MPH4; Lilith Tatham, DVM5; Janice Matthews-Greer, PhD4; Kevin Sohner, PhD5; Andrew S. Bowman, DVM, PhD6; James Avrill, DVM, PhD7; Tony Forshey, DVM8; Lenee Blanton, MPH2; C. Todd Davis, PhD2; John Schiltz, DVM9; Susan Skorupski, DVM10; LaShondra Berman, PhD2; Yunho Jang, PhD2; Joseph S. Bresee, MD2; Stephen Lindstrom, PhD2; Susan C. Trock, DVM2; David Wentworth, PhD2; Alicia M. Fry, MD2; Sietske de Fijter, MS5; Kimberly Signs, DVM4; Mary DiOrio, MD5; Sonja J. Olsen, PhD2; Matthew Biggerstaff, MPH2 (View author affiliations)
View suggested citationSummary
What is already known about this topic?
Sporadic human infections and outbreaks with influenza viruses that normally circulate in swine have occurred in the past. The largest known outbreak of H3N2v virus infections occurred in 2012.
What is added by this report?
In August 2016, 18 laboratory-confirmed infections with H3N2v virus were reported among persons who had attended agricultural fairs in Michigan and Ohio. Sixteen of the 18 cases occurred in persons who were infected with a reassortant H3N2v virus that contained a hemagglutinin (HA) gene previously not detected in variant viruses. The HA gene was likely introduced from humans into swine in 2010 or 2011, and viruses with this gene have circulated and evolved in swine to be genetically and antigenically different from both previous and currently circulating human seasonal influenza A(H3N2) viruses.
What are the implications for public health practice?
To minimize transmission of influenza viruses from swine to humans and from humans to swine, agricultural fair organizers should consider measures such as shortening the time swine are on the fairgrounds to ≤72 hours, immediately isolating ill swine, maintaining a veterinarian on call for the duration of the swine exhibition, providing prominent handwashing stations, and prohibiting food and beverages in animal barns. Persons at high risk for influenza-associated complications should be discouraged from entering swine barns.
On August 3, 2016, the Ohio Department of Health Laboratory reported to CDC that a respiratory specimen collected on July 28 from a male aged 13 years who attended an agricultural fair in Ohio during July 22–29, 2016, and subsequently developed a respiratory illness, tested positive by real-time reverse transcription–polymerase chain reaction (rRT-PCR) for influenza A(H3N2) variant* (H3N2v). The respiratory specimen was collected as part of routine influenza surveillance activities. The next day, CDC was notified of a child aged 9 years who was a swine exhibitor at an agricultural fair in Michigan who became ill on July 29, 2016, and tested positive for H3N2v virus at the Michigan Department of Health and Human Services Laboratory. Investigations by Michigan and Ohio health authorities identified 18 human infections linked to swine exhibits at agricultural fairs. To minimize transmission of influenza viruses from infected swine to visitors, agricultural fair organizers should consider prevention measures such as shortening the time swine are on the fairgrounds, isolating ill swine, maintaining a veterinarian on call, providing handwashing stations, and prohibiting food and beverages in animal barns. Persons at high risk for influenza-associated complications should be discouraged from entering swine barns.
Epidemiologic Investigation
Novel influenza viruses are different from currently circulating human influenza H1 and H3 viruses and have the potential to cause a pandemic if the virus is efficiently transmitted from person to person. In the United States, human infection with a novel influenza A virus is nationally notifiable, and globally, it is a reportable event under International Health Regulations 2005 (1); all such infections identified in the United States are investigated and reported to CDC. In early July 2016, before the identification of H3N2v virus infections described in this report, CDC reminded public health and laboratory partners nationwide to collect and test respiratory specimens from patients with influenza-like illness and swine exposure.
After the initial identification of the H3N2v virus infections, the Michigan Department of Health and Human Services and the Ohio Department of Health encouraged enhanced surveillance and increased collection and rRT-PCR testing of respiratory specimens from patients with illness and swine exposure or agricultural fair attendance. Specimens from swine at fairs in both states were tested for influenza by The Ohio State University in collaboration with the St. Jude Center of Excellence for Influenza Research and Surveillance and the United States Department of Agriculture-Animal and Plant Health Inspection Service-Veterinary Services, National Veterinary Services Laboratories, Influenza A Virus in Swine Surveillance Program.
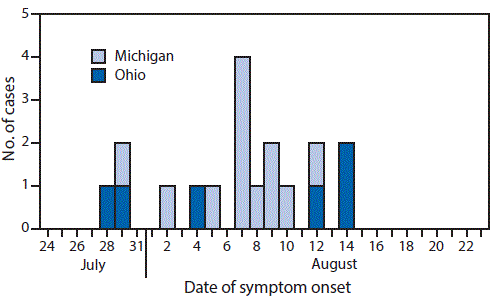
During August 3–25, 2016, a total of 18 human infections with H3N2v virus (12 from Michigan and six from Ohio) were confirmed and reported to CDC (Figure). All of the clinical specimens were sent to CDC for verification and further virus characterization. Sixteen of the ill persons were aged <18 years, including seven aged <5 years. All 18 persons reported exposure to swine during attendance at one or more of seven agricultural fairs (three in Michigan and four in Ohio); no ill person reported contact with another known infected person, and no person-to-person transmission was identified. Thirteen persons reported direct contact with swine (touching or handling), including four children aged <18 years who exhibited swine as part of a youth agriculture club. Four of the five persons who did not report direct swine contact reported passing through a swine barn; the fifth person was a fair attendee with unspecified indirect contact with swine. Specimens obtained from swine from all seven fairs tested positive for influenza A(H3N2) virus. Eight of the 18 ill persons were at high risk for influenza-associated complications because of the presence of an underlying medical condition or because of their young age (2). One person, who had an underlying condition, was hospitalized for 2 days. All persons fully recovered. Six ill persons were treated with an influenza antiviral medication. Among the 17 persons with known vaccination history, three had received a seasonal influenza vaccination in the preceding 12 months.
Laboratory Investigation
CDC performed genomic sequence analysis on all 18 specimens from infected humans; two different H3N2v viruses were identified. Among the 18 variant viruses detected in persons, 16 were reassortants, with a constellation of genes not previously detected in viruses infecting humans. Whereas these viruses contained seven gene segments similar to segments detected in previously reported variant virus outbreaks, one gene segment coding for an influenza A(H3) hemagglutinin (HA) gene was determined to be similar to HA genes found in human seasonal influenza A(H3N2) viruses from 2010 and 2011. This HA gene was likely introduced from humans into swine in 2010 or 2011, and has since circulated and evolved in swine to be genetically and antigenically different from both previous and currently circulating human seasonal influenza A(H3N2) viruses (3). The viruses in the remaining two specimens had HA genes similar to those of swine-origin H3N2 influenza viruses circulating in the U.S. swine population since 1998, and previously identified in human H3N2v virus infections in the United States since 2009 (4,5). Preliminary analysis suggests that the viruses identified in all human specimens were nearly identical to H3N2 viruses detected in swine at agricultural fairs in Michigan and Ohio. With the exception of one virus isolated from a human specimen, H3N2 viruses found in human cases were genetically related to specimens from swine at the same fairs attended by the infected persons.
All fully sequenced viruses from human infections had the matrix (M) gene from the influenza A(H1N1)pdm09 virus, which has been seen in H3N2v outbreaks since 2011 and has been found to enhance transmissibility in several animal models (6,7) and confer resistance to adamantine antiviral drugs (amantadine and rimantadine). Four H3N2v viruses were tested and were susceptible to neuraminidase inhibitors (oseltamivir, peramivir, and zanamivir).
Discussion
Including the infections described in this report, 372 H3N2v virus infections have been reported in the United States since human infections with novel influenza A viruses became notifiable in 2005 (8); this report describes all reported H3N2v virus infections in the United States since January 1, 2016. This outbreak underscores the importance of implementing measures to minimize influenza transmission between swine and persons at agricultural fairs. Young persons who exhibit or have other direct contact with swine should know how to protect themselves from infection, as most infections in this and previous outbreaks have been in persons aged <18 years who had direct contact with swine (5). Transmission is possible, though less common, without direct swine contact. In this outbreak, five of the 18 infections occurred in persons who did not report direct contact with swine. Messaging about variant influenza virus transmission risk and prevention, particularly among young persons and persons at increased risk for influenza-associated complications (children aged <5 years, persons aged ≥65 years, pregnant women, and persons with certain health conditions) (2), should be provided to all agricultural fair organizers and officials, fair attendees, and animal exhibitors. When planning for subsequent fair seasons, fair organizers should consider prevention measures such as shortening the time swine are on the fairgrounds to ≤72 hours (9), establishing a protocol to immediately isolate ill swine, maintaining a veterinarian on call for the duration of the swine exhibition, providing prominent handwashing stations in or near animal barns for exhibitors and attendees, and displaying signage that discourages or prohibits food and beverages in animal barns (9,10) and discourages persons at high risk for influenza-associated complications from entering swine barns.
Treatment with a neuraminidase inhibitor is recommended for persons with suspected variant virus infection who are hospitalized, who have severe or progressive illness, or who are in a group at high risk for influenza-associated complications. Treatment with these influenza antiviral drugs also can be considered for any previously healthy outpatient with confirmed or suspected H3N2v virus infection based on clinical judgment. Human seasonal influenza vaccine is not known to protect against commonly circulating swine-origin influenza viruses, but it can protect against seasonal influenza, which can circulate even during summer months when most fairs occur. An annual seasonal influenza vaccine, which is recommended for all persons aged ≥6 months, might help prevent future reassortment of H3N2v viruses with human seasonal influenza viruses.
Rapid detection and reporting of human infections with novel influenza A viruses are important to facilitate prompt identification and characterization of influenza A viruses with pandemic potential and to accelerate the implementation of an effective public health response. Most persons in this outbreak were infected with an influenza virus genotype not previously detected in humans. Although person-to-person spread was not identified, ongoing investigations to monitor for genetic changes in the virus and to detect person-to-person transmission continue to be necessary. Health care providers should consider novel influenza virus infections in ill persons with swine exposure or agricultural fair attendance and consult with their state public health department about further testing, regardless of the results of rapid influenza diagnostic tests (which can have poor sensitivity for variant viruses), based on CDC’s interim guidance for clinicians.† All clinical specimens that are suspected to contain novel influenza A viruses should be sent by state public health laboratories to CDC for additional characterization and verification of results. Additional information about variant influenza viruses is available at http://www.cdc.gov/flu/swineflu/h3n2v-cases.htm.
Acknowledgments
Susan Bohm, Mary Grace Stobierski, Laura Mosher, Kevin Rodeman, Michigan Department of Health and Human Services; Alicia Janas-Martindale, Mary Lea Killian, U.S. Department of Agriculture; Jacqueline Nolting, Ohio State University; Steve Hussey, Michigan Department of Agriculture and Rural Development; Julie Thelen, Michigan State University; Yan Zhang, Ohio Department of Agriculture; staff members in county health departments in Michigan and Ohio.
Corresponding author: Rebekah S. Schicker, rschicker@cdc.gov, 404-639-3747.
1Epidemic Intelligence Service, CDC; 2Influenza Division, National Center for Immunization and Respiratory Diseases, CDC; 3Epidemiology Elective Program, Division of Scientific Education and Professional Development, Center for Surveillance, Epidemiology, and Laboratory Services, CDC; 4Michigan Department of Health and Human Services; 5Ohio Department of Health; 6College of Veterinary Medicine, Ohio State University; 7Animal Industry Division, Michigan Department of Agricultural and Rural Development; 8Division of Animal Health, Ohio Department of Agriculture; 9National Veterinary Services Laboratories, U.S. Department of Agriculture; 10Animal and Plant Health Inspection Service, U.S. Department of Agriculture.
* Influenza viruses that normally circulate in swine are referred to as “variant” when they infect humans. Seasonal influenza A(H3N2) viruses that circulate worldwide in the human population have substantial antigenic and genetic differences from influenza A(H3N2) viruses circulating in swine (http://www.who.int/influenza/gisrs_laboratory/terminology_variant/en/).
†http://www.cdc.gov/flu/swineflu/interim-guidance-variant-flu.htm.
References
- Council of State and Territorial Epidemiologists. National reporting for initial detections of novel influenza A viruses. Position statement 07–ID01. Atlanta, GA: Council of State and Territorial Epidemiologists; 2007. http://c.ymcdn.com/sites/www.cste.org/resource/resmgr/PS/07-ID-01.pdf
- CDC. Influenza (Flu): influenza A (H3N2) variant virus. CDC recommendations for people at high risk. Atlanta, GA: CDC; 2016. http://www.cdc.gov/flu/swineflu/h3n2v-cases.htm
- Rajão DS, Gauger PC, Anderson TK, et al. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol 2015;89:11213–22. CrossRef PubMed
- Epperson S, Jhung M, Richards S, et al. ; Influenza A (H3N2)v Virus Investigation Team. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin Infect Dis 2013;57(Suppl 1):S4–11. CrossRef PubMed
- Jhung MA, Epperson S, Biggerstaff M, et al. Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis 2013;57:1703–12. CrossRef PubMed
- Chou YY, Albrecht RA, Pica N, et al. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol 2011;85:11235–41. CrossRef PubMed
- Ma J, Shen H, Liu Q, et al. Pathogenicity and transmissibility of novel reassortant H3N2 influenza viruses with 2009 pandemic H1N1 genes in pigs. J Virol 2015;89:2831–41. CrossRef PubMed
- CDC. Influenza (Flu): reported infections with variant influenza viruses in the United States since 2005. Atlanta, GA: CDC; 2016. http://www.cdc.gov/flu/swineflu/variant-cases-us.htm
- National Assembly of State Animal Health Officials; National Association of State Public Health Veterinarians. Measures to minimize influenza transmission at swine exhibitions. Arlington, VA: National Assembly of State Animal Health Officials; 2014. https://www.cdc.gov/flu/pdf/swineflu/influenza-transmission-swine-exhibitions-2014.pdf
- Bowman AS, Workman JD, Nolting JM, Nelson SW, Slemons RD. Exploration of risk factors contributing to the presence of influenza A virus in swine at agricultural fairs. Emerg Microbes Infect 2014;3:e5. CrossRef PubMed
 FIGURE. Influenza A(H3N2) variant virus infections (N = 18), by date of symptom onset — Michigan and Ohio, July–August 2016
FIGURE. Influenza A(H3N2) variant virus infections (N = 18), by date of symptom onset — Michigan and Ohio, July–August 2016
Suggested citation for this article: Schicker RS, Rossow J, Eckel S, et al. Outbreak of Influenza A(H3N2) Variant Virus Infections Among Persons Attending Agricultural Fairs Housing Infected Swine — Michigan and Ohio, July–August 2016. MMWR Morb Mortal Wkly Rep 2016;65:1157–1160. DOI: http://dx.doi.org/10.15585/mmwr.mm6542a1.
Use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of
Health and Human Services.
References to non-CDC sites on the Internet are
provided as a service to MMWR readers and do not constitute or imply
endorsement of these organizations or their programs by CDC or the U.S.
Department of Health and Human Services. CDC is not responsible for the content
of pages found at these sites. URL addresses listed in MMWR were current as of
the date of publication.
All HTML versions of MMWR articles are generated from final proofs through an automated process. This conversion might result in character translation or format errors in the HTML version. Users are referred to the electronic PDF version (https://www.cdc.gov/mmwr) and/or the original MMWR paper copy for printable versions of official text, figures, and tables.
Questions or messages regarding errors in formatting should be addressed to mmwrq@cdc.gov.
- Page last reviewed: August 17, 2017
- Page last updated: August 17, 2017
- Content source:


 ShareCompartir
ShareCompartir