Kingella denitrificans
Introduction
K. denitrificans is one of several species which are colistin-resistant and may be routinely isolated on selective medium for N. gonorrhoeae. Colonies of K. denitrificans are usually glistening and smooth in consistency. Although cells of K. denitrificans are coccobacilli, the cell morphology of some isolates may appear to be diplococci and very similar in appearance to N. gonorrhoeae.
In contrast to Neisseria spp., K. denitrificans isolates are catalase- and superoxol-negative. N. gonorrhoeae and K. denitrificans may, however, be confused because isolates of both species produce acid from glucose in acid production tests and hydroxylprolylaminopeptidase in enzyme substrate tests. Crossreactions also have been noted with isolates of K. denitrificans in coagglutination serologic tests for the identification of N. gonorrhoeae. Thus, additional tests must be performed to differentiate between these species.
The nitrate test permits differentiation between N. gonorrhoeae (nitrate-negative) and K. denitrificans (nitrate-positive).
Characteristics of K. denitrificans
Table 1. Characteristics of K. denitrificans
| Characteristic | Illustration |
|---|---|
| Gram stain Cell Morphology | Gram-negative coccobacilli/rods |
Colonies of K. denitrificans are quite opaque and tend to adhere to the agar surface, particularly after incubation for 48 h. | 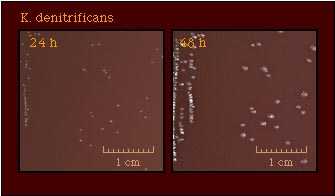 |
| Pigmentation | 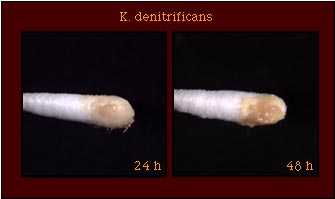 |
| Oxidase Test | 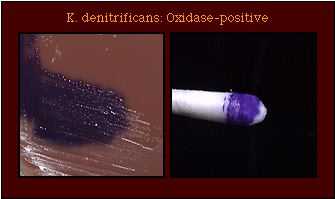 |
| Acid Production |  Acid only from glucose |
| Enzyme Substrate Test | Prolylaminopeptidase-positive |
| Nitrate Reduction Test | Nitrate +ve |
| Polysaccharide from Sucrose |  Polysaccharide -ve |
| Production of Deoxyribonuclease (DNase) |  DNase -ve |
| Superoxol Test (Reaction with 30% hydrogen peroxide) |  Superoxol -ve |
| Catalase Test (Reaction with 3% hydrogen peroxide) |  Catalase -ve |
| Colistin Resistance |  Colistin-resistant |
Species which may be misidentified as K. denitrificans in acid detection tests
Depending on the identification test used, several strains of Neisseria may be misidentified as K. denitrificans; the biochemical characteristics of these species are shown in Table 2.
Table 2. Supplemental tests which permit differentiation among gram-negative diplococci that produce acid only from glucose.
| Species that produce acid only from glucose | Gram Stain | Enzyme substrate | Nitrate Reduction | DNase | Colistin Resistance |
|---|---|---|---|---|---|
| K. denitrificans | GNR* | Hydroxy- prolylaminopeptidase +ve | + | - | R |
| N. gonorrhoeae | GND | Hydroxy- prolylaminopeptidase +ve | - | - | R |
| Maltose -ve N. meningitidis | GND | Gamma-glutamyl- aminopeptidase +ve | - | - | R |
| "N. kochii" | GND | Hydroxy- prolylaminopeptidase +ve | - | - | R |
| N. cinerea** | GND | Hydroxy- prolylaminopeptidase +ve | - | - | (R) |
| N. elongata** | GNR | Hydroxy- prolylaminopeptidase +ve | + | + | (R) |
| M. catarrhalis** | GND | No reaction | + | + | (R) |
Abbreviations. GND, Gram-negative diplococcus; GNR, Gram-negative rod; +, most strains positive; -, most strains negative; R, strains grow well on selective medium for N. gonorrhoeae and/or show no inhibition around a colistin disk (10 micrograms); (R), most strains susceptible, some strains resistant.
*Cellular elongation test will allow differentiation between rods and cocci.
**Some strains of N. cinerea, N. elongata, and M. catarrhalis give weak positive reactions although this characteristic is not representative of the species.
Although enzyme substrate tests are intended to be used only for the identification of Neisseria spp. isolated on selective media for N. gonorrhoeae, isolates of K. denitrificans give the same reaction (hydroxylaminopeptidase-positive) in this test as do isolates of N. gonorrhoeae. Thus additional tests must be performed to differentiate between these species.
Table 3. Supplemental tests which permit differentiation among Neisseria and related species that produce hydroxyprolylaminopeptidase.
| Species that Produce Hydroxylprolylamino- peptidase | Cellular Elongation* | Acid from | Nitrate Reduction | Polysaccharide from Sucrose | Superoxol | Colistin Resistance | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| G | M | S | F | L | ||||||
| N. polysaccharea | Diplococci | + | - | - | - | - | - | - | Weak (4+) positive | (R) |
| K. denitrificans | Rod filaments | + | - | - | - | - | + | - | - | R |
| N. gonorrhoeae "N. kochii"* | Diplococci | + | - | - | - | - | - | - | Strong (4+) positive | R |
| N. cinerea | Diplococci | - | - | - | - | - | - | - | Weak (2+) positive | (R) |
| N. subflava biovar subflava | Diplococci | + | + | - | - | - | - | - | Weak (2+) positive | S |
| N. subflava biovar flava | Diplococci | + | + | - | + | - | - | - | Weak (2+) positive | S |
| N. subflava biovar perflava | Diplococci | + | + | + | + | - | - | + | Weak (2+) positive | (R) |
| N. sicca | Diplococci | + | + | + | + | - | - | + | Weak (2+) positive | S |
| N. mucosa | Diplococci | + | + | + | + | - | + | + | Weak (2+) positive | S |
| N. flavescens | Diplococci | - | - | - | - | - | - | + | Weak (2+) positive | R |
| N. elongata | Rod filaments | (+) | - | - | - | - | - | - | - | (R) |
Abbreviations: +, most strains positive; -, most strains negative; (+), some strains give weak positive reactions which are not representative of the species; R, strains grow well on selective medium for N. gonorrhoeae and/or show no inhibition around a colistin disk (10 micrograms); (R), most strains susceptible, some strains resistant.
References
Bovre K. 1984. Family VIII. Neisseriaceae Prevot, p. 288-309. In N. R. Krieg (ed.). Manual of Systematic Bacteriology, vol. 1. The Williams & Wilkins co., Baltimore.
Knapp JS. Historical perspectives and identification of Neisseria and related species. Clin Microbiol Rev 1988;1:415-431.
Knapp JS, Rice RJ. Neisseria and Branhamella. In. Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH. (ed.). Manual of Clinical Microbiology. 6th ed. American Society for Microbiology, Washington D. C, 1995.
Vedros NA. 1984. Genus I. Neisseria Trevisan 1885, 105AL, p. 290-296. In NR Krieg (ed.). Bergey's Manual of Systematic Bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore.
- Page last reviewed: March 31, 2017
- Page last updated: October 17, 2008
- Content source:


 ShareCompartir
ShareCompartir