2016 STD Prevention Conference Highlights
This web page is archived for historical purposes and is no longer being updated.
Antibiotic-Resistant Gonorrhea
The 2016 STD Prevention Conference showcases the latest STD prevention research, both from the United States and abroad.
This page highlights one presentation and one scientific poster with a focus on antibiotic-resistant gonorrhea, including new testing technology to monitor for resistance and an analysis of the medical costs that can be averted by keeping the prevalence of resistance low.
Poster or presentation images, as well as full abstracts and related links follow below.
Fast Facts
- Of new gonorrhea infections, 30% are resistant to at least 1 drug.
- Gonorrhea is the second most commonly reported communicable disease in the U.S., with an estimated 820,000 new infections each year.
- In 2006, CDC recommended five treatment options for gonorrhea – we now have only one.
Antibiotic-Resistant Gonorrhea Abstracts
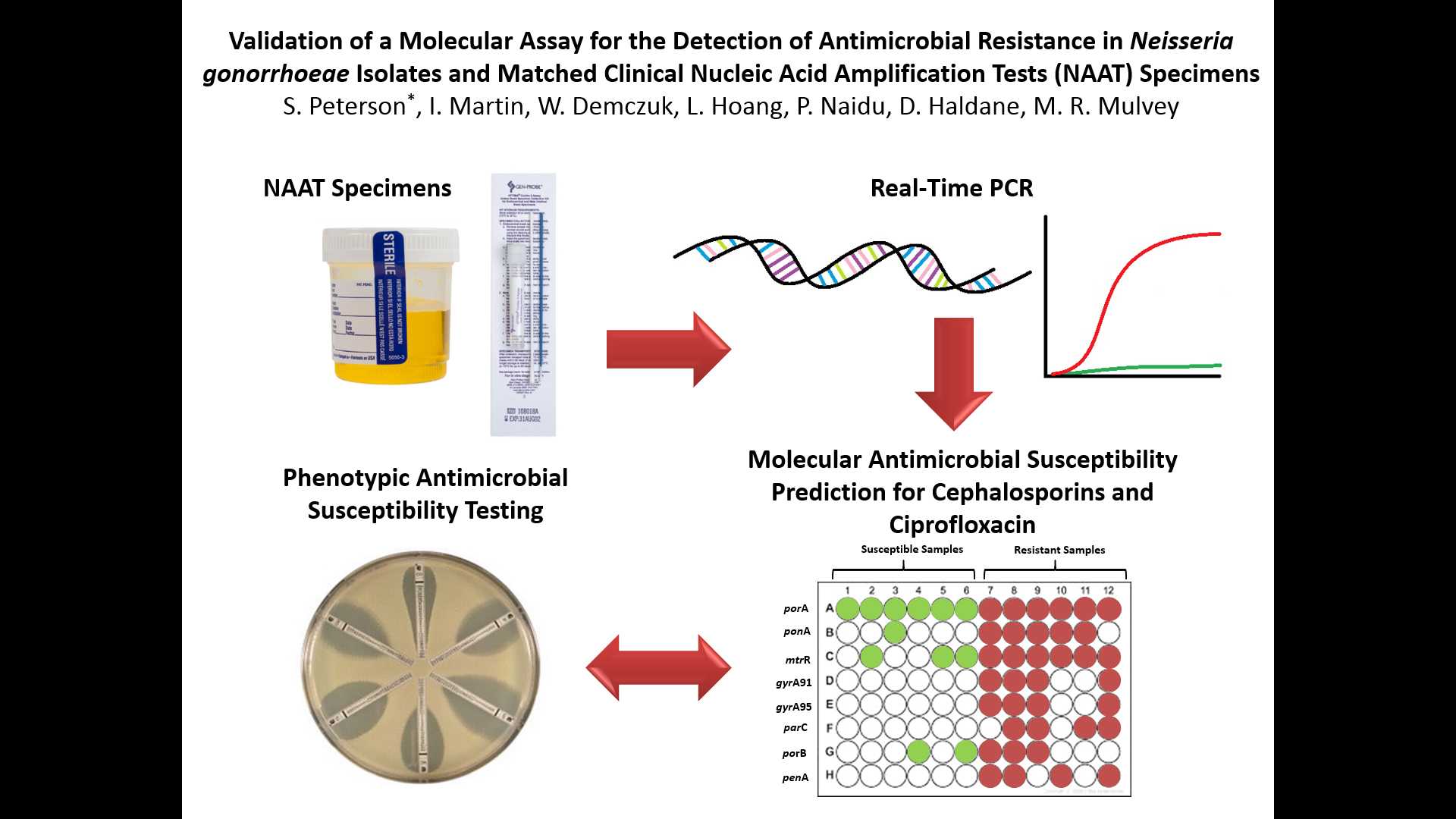
VALIDATION OF A MOLECULAR ASSAY FOR THE DETECTION OF ANTIMICROBIAL RESISTANCE IN NEISSERIA GONORRHOEAE ISOLATES AND MATCHED CLINICAL NUCLEIC ACID AMPLIFICATION TESTS (NAAT) SPECIMENS
S. Peterson1*, I. Martin1, W. Demczuk1, L. Hoang2, P. Naidu3, D. Haldane4, M. R. Mulvey1
1Bacteriology and Enteric Diseases Program, National Microbiology Laboratory, Public Health Agency of Canada
2British Columbia Centres for Disease Control Public Health Microbiology & Reference Laboratory, Vancouver, British Columbia
3Provincial Laboratory for Public Health, Edmonton, Alberta
4Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia
Background: The incidence of antimicrobial resistant Neisseria gonorrhoeae continues to rise in Canada. Due to increased Nucleic Acid Amplification Testing (NAAT) for gonorrhea diagnosis, antimicrobial resistance data on ~70% of gonorrhea infections diagnosed by NAAT yearly is not available. A molecular method was developed to monitor antimicrobial resistance in N. gonorrhoeae using bacterial cultures and NAAT specimens.
Method: N. gonorrhoeae multi antigen sequence type (NG-MAST) was determined and real-time (RT-) PCR assays were developed to detect single nucleotide polymorphisms (SNPs) in seven genes associated with extended-spectrum cephalosporin (ESC), ciprofloxacin (CIP), or azithromycin (AZR) resistance (ponA, mtrR, penA, gyrA, parC, porB, 23S rRNA) in 112 clinical NAAT specimens (89 APTIMA, 6 BD Viper, 4 neat urine, 13 Roche-Cobas) and matching bacterial cultures. RT-PCR results were compared to sequences determined by whole genome sequencing (WGS) and minimum inhibitory concentrations (MIC) for ESC and CIP were determined by agar dilution.
Results: SNP assay analysis and NG-MAST were successfully determined for 101 of 112 matched specimens. Decreased (MIC≥0.125µg/ml), moderate (MIC=0.032-0.063µg/ml), and full (MIC<0.032µg/ml) susceptibility to ESC (based on the number of SNPs) was observed in 6.9% (n=7), 15.8% (n=16) and 72.3% (n=73) of NAAT specimens, respectively, (5% (n=5) undetermined); and 5% (n=5), 25.7% (n=26) and 69.3% (n=70) of the bacterial cultures. CIP resistance was predicted in 24.7% (n=25) of NAAT specimens and 67.3% (n=68) were predicted susceptible, with 7.9% (n=8) undetermined; whereas 23.8% (n=24) of cultures were resistant and 76.2% (n=77) were susceptible. Agreement of SNPs detected between successful RT-PCR assays and WGS was 100% for all SNP assays.
Conclusions: The utility of a RT-PCR assay for detection of known antimicrobial resistance markers was demonstrated to be useful to enhance surveillance by estimating antimicrobial resistance in situations where bacterial cultures of N. gonorrhoeae are no longer available.
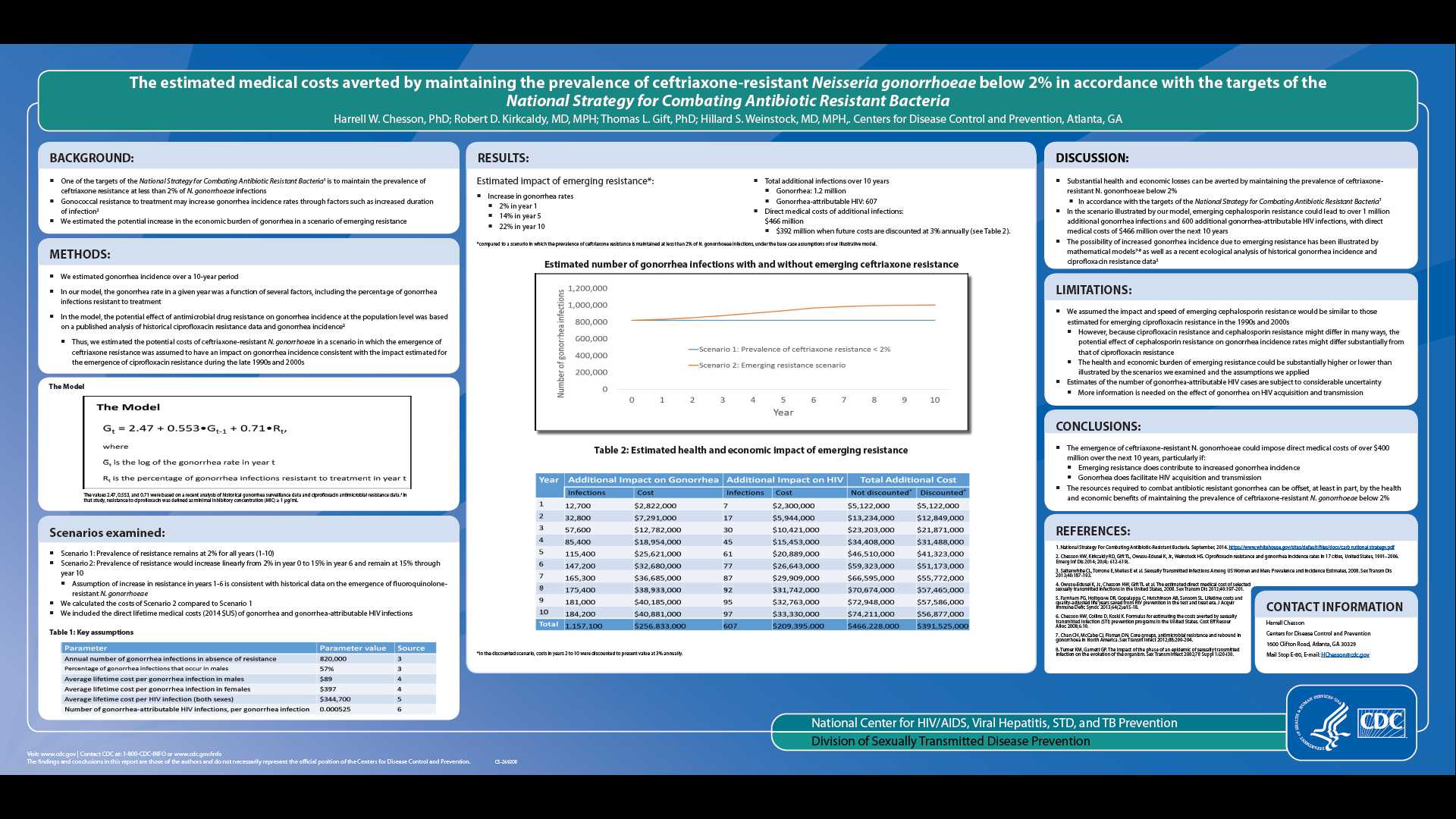
THE ESTIMATED MEDICAL COSTS AVERTED BY MAINTAINING THE PREVALENCE OF CEFTRIAXONE-RESISTANT NEISSERIA GONORRHOEAE BELOW 2% IN ACCORDANCE WITH THE TARGETS OF THE NATIONAL STRATEGY FOR COMBATING ANTIBIOTIC RESISTANT BACTERIA
Harrell Chesson, PhD, Bob Kirkcaldy, MD, MPH, Thomas Gift, PhD and Hillard S. Weinstock, MD, MPH
Division of STD Prevention, Centers for Disease Control and Prevention, Atlanta
Background: One of the targets of the National Strategy for Combating Antibiotic Resistant Bacteria is to maintain the prevalence of ceftriaxone-resistant Neisseria gonorrhoeae at less than 2%. Gonococcal resistance can potentially increase gonorrhea incidence rates through factors such as increased duration of infection. We estimated the potential increase in the economic burden of gonorrhea in a scenario of emerging resistance.
Methods: We estimated gonorrhea incidence over a ten year period (year 1 to year 10) using a simple model in which the gonorrhea rate in a given year was a function of several factors, including the percentage of gonorrhea cases resistant to treatment in the previous year. Model assumptions were informed by a published analysis of historical gonorrhea surveillance data and antimicrobial resistance data. We assumed 820,000 cases of gonorrhea annually in the absence of resistance. In the first scenario, we assumed the prevalence of resistance would remain at 2% for years 1-10. In the second scenario, we assumed the prevalence of resistance would increase linearly from 2% in year 1 to 15% in year 6, as observed during the emergence of fluoroquinolone-resistant N. gonorrhoeae, and remain at 15% through year 10. We included the direct lifetime medical costs (2014 $US) of gonorrhea and gonorrhea-attributable HIV cases, based on published cost and probability estimates.
Results: Over 10 years and when compared to maintaining prevalence of resistance at 2%, emerging resistance would contribute to an estimated 1,157,000 additional gonorrhea cases (costing $257 million) and 607 additional gonorrhea-attributable HIV cases (costing $209 million), for a total cost of $466 million without discounting and $392 million when future costs are discounted at 3% annually.
Conclusions: Preventing the emergence of ceftriaxone-resistant N. gonorrhoeae in accordance with the National Strategy for Combating Antibiotic Resistant Bacteria targets can potentially yield substantial health and economic benefits.
- Page last reviewed: September 7, 2016 (archived document)
- Content source:


 ShareCompartir
ShareCompartir