Antibiotic Resistance Lab Network
About the AR Lab Network
Established in 2016, CDC’s Antibiotic Resistance Laboratory Network (AR Lab Network) supports nationwide lab capacity to rapidly detect antibiotic resistance in healthcare, food, and the community, and inform local responses to prevent spread and protect people. The AR Lab Network includes seven regional labs, the National Tuberculosis Molecular Surveillance Center (National TB Center), and labs in 50 states, six cities, and Puerto Rico. As a whole, the network tracks changes in resistance and helps identify and respond to outbreaks faster.
AR Lab Network Regional Labs and the National TB Molecular Surveillance Center detect existing and emerging types of antibiotic resistance, track emerging resistance more effectively, and generate stronger data to protect people and combat future resistance threats. This ambitious approach transforms the nation’s laboratory landscape by establishing the much-needed infrastructure across the nation. This infrastructure will allow the public health community to rapidly detect emerging AR threats, sound the alarm for a comprehensive local response, and better understand these deadly threats so we can contain them quickly.
All 50 states, six large cities, and Puerto Rico received investments in 2016 and 2017 to increase capabilities to test for carbapenem-resistant Enterobacteriaceae (CRE) and carbapenem-resistant Pseudomonas aeruginosa (CRPA) and perform whole genome sequencing (WGS) on all Salmonella isolates. The Salmonella testing builds on the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS), a public health surveillance system. The regional labs will complement these activities done at the local and state level by providing additional or confirmation testing and outbreak support for these pathogens, when needed.
The AR Lab Network complements additional CDC AR prevention and response investments, collectively known as CDC’s AR Solutions Initiative, to amplify prevention and response to fight antibiotic resistance nationwide.
How the AR Lab Network Works
When new resistance threats or outbreaks are detected within healthcare facilities or state and local labs, regional labs provide support, where needed, to characterize, support response, and track these discoveries. Since outbreak response varies by state, the support launched by the AR Lab Network may also vary by state or threat discovered. The regional labs will ensure more consistent and improved communication, coordination, and tracking at all levels—across healthcare facilities, state health departments, and CDC—every time.
The regional labs and National TB Center work together with CDC and state and local health department labs to:
- Detect new resistance and provide better big-picture trend tracking to create pathogen-specific solutions and support national public health strategies.
- Inform outbreak response when AR threats, like CRE, are reported, working together with state and local labs.
- Prevent and combat future AR threats by creating better data.
- Support innovations in antibiotic and diagnostic development. Samples from the labs will be made available through the CDC and FDA AR Isolate Bank, which researchers can use to develop earlier diagnoses and more effective treatment options.
The AR Lab Network Regional Labs also conduct targeted surveillance, as requested, for urgent, serious, and concerning threats identified by CDC, like Clostridium difficile, vancomycin-resistant Staphylococcus aureus (VRSA), and vancomycin-resistant enterococci (VRE).
About the AR Lab Network Regional Labs and National TB Molecular Surveillance Center
The regional labs and National TB Center were strategically placed in regions of the United States to help track changes in resistance and help identify outbreaks with local technical support. The regional labs coordinate local efforts in the state health departments in Maryland, Michigan, Minnesota, New York, Tennessee, Texas, Washington, and Wisconsin. Regions are defined by an existing regionalization scheme used by PulseNet.
On this page:

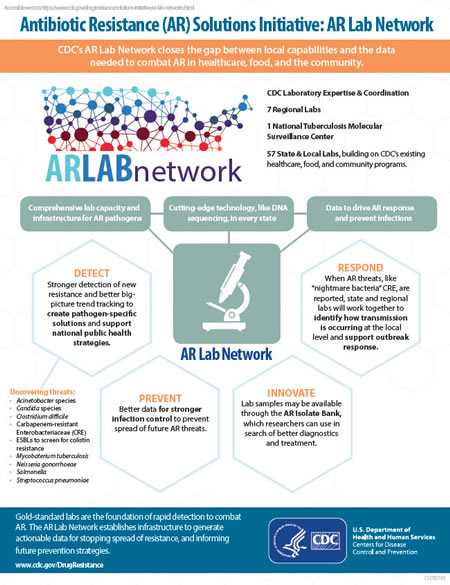
The AR Lab Network closes the gap between capabilities and data needed to combat AR.
- Comprehensive lab capacity and infrastructure for AR pathogens.
- Cutting-edge technology, like DNA sequencing, in every state.
- Data to drive AR response and prevent infections
Detect: Stronger detection of new resistance and better big picture trend tracking to create pathogen-specific solutions and support national public health strategies. Uncovering threats:
- Acinetobacter species
- Candida species
- Clostridium difficile
- Carbapenem-resistant
- Enterobacteriaceae (CRE)
- ESBLs to screen for colistin resistance
- Mycobaterium tuberculosis
- Neisseria gonorrhoeae
- Salmonella
- Streptococcus pneumoniae
Prevent: Better data for stronger infection control to prevent spread of future AR threats.
Innovate: Lab samples may be available through the AR Isolate Bank, which researchers can use in search of better diagnostics and treatment.
Respond: When AR threats, like “nightmare bacteria” CRE, are reported, state and regional labs will work together to identify how transmission is occurring at the local level and support outbreak response.
Gold-standard labs are the foundation of rapid detection to combat AR.
Every regional lab does core testing. Select labs provide additional testing to support nationwide needs. The national tuberculosis center is equipped to perform WGS for isolates of Mycobacterium tuberculosis in the U.S.
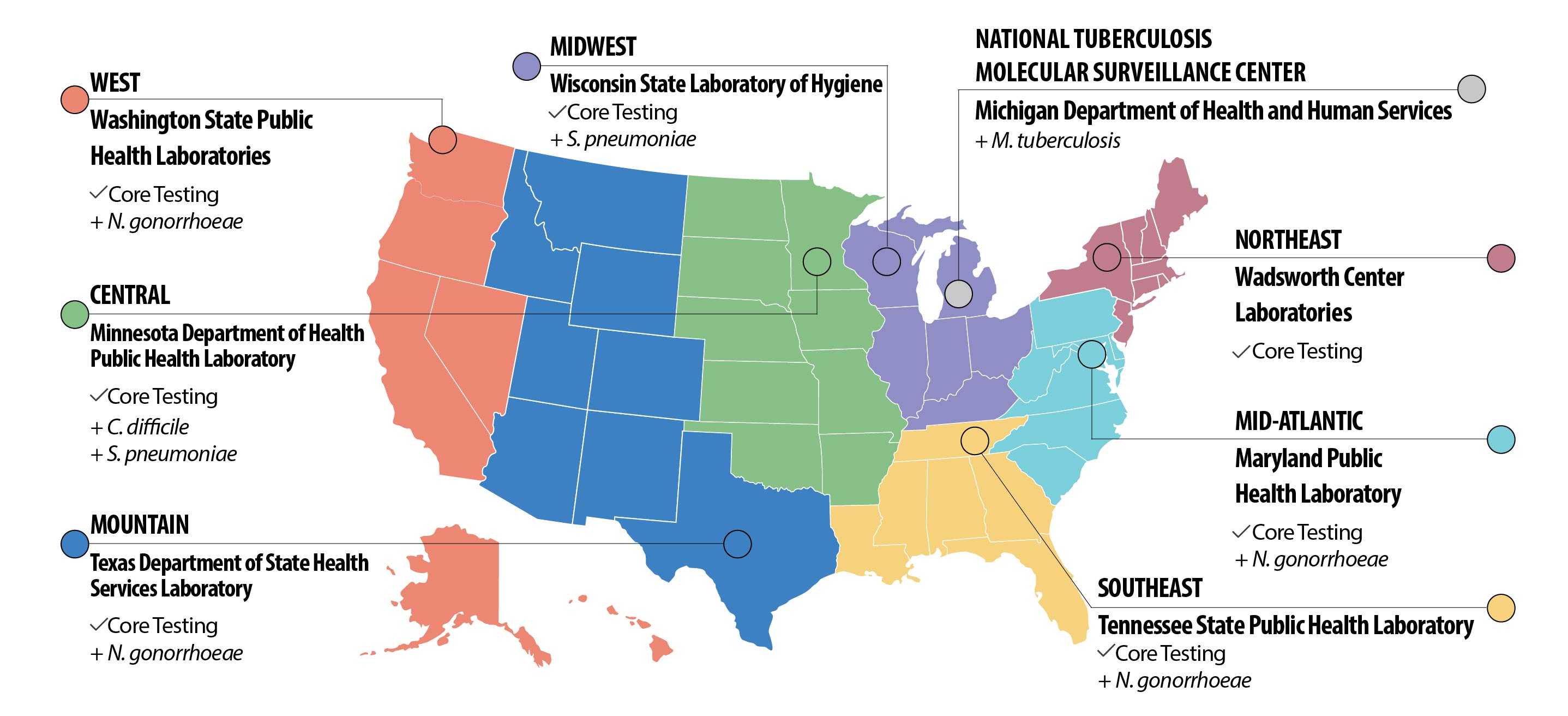
Infographic Details
Antibiotic Resistance (AR) Lab Network Regional Labs Map
Midwest
Wisconsin State Laboratory of Hygiene
- Core Testing
+ pneumoniae
National Tuberculosis Molecular Surveillance Center
- Michigan Department of Health and Human Services
+tuberculosis
Northeast
Wadsworth Center Laboratories
- Core Testing
Mid-Atlantic
Maryland Public Health Laboratory
- Core Testing
+ gonorrhoeae
Southeast
Tennessee State Public Health Laboratory
- Core Testing
+ gonorrhoeae
Mountain
Texas Department of State Health Services Laboratory
- Core Testing
+ gonorrhoeae
Central
Minnesota Department of Health Public Health Laboratory
- Core Testing
+ difficile
+ pneumoniae
West
Washington State Public Health Laboratories
- Core Testing
+ gonorrhoeae
All regional labs will perform Core Testing for their region, including:
- Molecular testing to detect colonization of carbapenem-resistant Enterobacteriaceae (CRE).
- Detection of new and emerging threats, like mcr-1, and ability to detect changes to known threats, like Methicillin-resistant Staphylococcus aureus (MRSA).
- Isolates may be used for the CDC and FDA AR Isolate Bank and WGS projects.
- Fungal susceptibility of Candida species to identify emerging resistance.
- Identification and colonization screening to detect and help prevent spread of Candida auris (C. auris).
Select labs will provide Additional Testing to support nationwide needs, including:
- Clostridium difficile special projects.
- Enhanced detection of drug-resistant Neisseria gonorrhea using WGS.
- Antimicrobial susceptibility and serotyping of multidrug-resistant Streptococcus pneumonia.
- Perform WGS for all isolates of Mycobacterium tuberculosis.
The Core and Additional Testing complements lab capacity supported by CDC in every state, six cities, and Puerto Rico.
For more information about the AR Lab Network, email ARLN@cdc.gov.
AR Lab Network Factsheets
- Page last reviewed: August 9, 2017
- Page last updated: September 20, 2017
- Content source:


 ShareCompartir
ShareCompartir
